Research Summary:
The goal of my research program over the past three decades has been to understand the contribution of transposable elements (TEs) to gene and genome evolution. TEs are fragments of DNA that can insert into new chromosomal locations and often make duplicate copies of themselves in the process. The analysis of genome sequences from both plants and animals has led to the surprising finding that TEs comprise the single largest component. They account for at least 45% of the human genome and 50-80% of some important plant genomes including maize, wheat and barley.
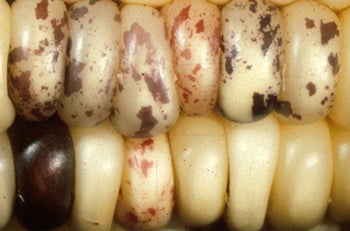
TEs were discovered in maize (corn) by Barbara McClintock over 50 years ago as the genetic agents that are responsible for the spots of pigmentation on mutant kernels. They are also responsible for the sectors of pigment in the rose.
As a postdoctoral fellow, I was involved in the isolation of the maize Ac and Ds elements [1]. My laboratory went on to discover that insertion of TEs altered maize gene expression in several interesting ways[2]
Computational analysis of transposable elements in plant genomes
In recent years we have pioneered the use of computational analysis (bioinformatics) in the identification and characterization of TEs in genome sequence. We have developed software including TARGeT [3] and MITE-Hunter [4] and used these and other programs to annotate TEs in the sequenced genomes of many plants including black cottonwood (Populus trichocarpa)[5], maize [6], rice [7], [8], Lotus japonicus [9], arabidopsis and Brassica oleracea [10], and mimulus and amborella (in preparation).
From the computer to the lab bench: developing transposition assays to find active TEs
Although most of the genomes of higher eukaryotes are derived from TEs, the vast majority can no longer transpose due to inactivating mutations or epigenetic silencing by the host. To analyze the role of TEs in gene and genome evolution it is important to identify TEs that are still active so that we can determine the impact of their movement on their host. To this end we first use computer programs to identify TEs that have features consistent with recent activity. Next, active candidates are tested using transposition assays we developed in the model organisms yeast [11], [12] and Arabidopsis [13]. The picture below shows a rice TE excising from a GFP reporter gene in transgenic Arabidopsis.
Studying the dynamic genome in real time: explosive amplification of a rice TE (funded by the NSF plant genome program)
Almost 20 years ago, members of my laboratory discovered MITEs (miniature inverted repeat transposable elements), a type of TE that is prevalent in the noncoding regions of plant genes[14], [15], [16]. Subsequently, MITEs were found to be abundant in the genes of several animal species, especially mosquitoes (more on this below). In order to determine the impact of MITE insertion on gene expression, we (in collaboration with the laboratory of Sean Eddy) used a novel computational strategy to identify an actively transposing MITE in rice [17] that was called mPing. In collaboration with the laboratory of Okumoto and Tanisaka (Kyoto University), we identified rice strains where mPing was increasing its copy number by over 40 new insertions per plant per generation [18]. Recently we used next generation high throughput sequencing to identify the new insertions in these strains and show that mPing can successfully amplify throughout the rice genome because most of the insertions have no impact on gene expression. Furthermore we showed that many mPing insertions make the adjacent rice gene transcription stress-inducible [19].
Are there active MITEs in mosquito genomes? (funded by the WM Keck Foundation)
We are collaborating with UC Riverside professors, Peter Atkinson (Entomology) and Jason Stajich (Plant Pathology and Microbiology) to characterize the MITEs in two recently sequenced mosquito genomes. To date investigators have not been able to engineer active elements from other organisms into tools for mosquito gene discovery. Our plan is to find active mosquito MITEs that could ultimately be exploited for this purpose. We will first use computational analysis to annotate all MITEs and element-encoding transposases in the genomes of Culex quinquefasciatus (a vector of West Nile virus) and Aedes aegypti (a vector for yellow and dengue fever viruses). We will then use a novel strategy involving a yeast transposition assay that recently proved highly effective at finding active MITEs and their transposase sources from rice genome sequence. By analyzing the results of over 1000 yeast crosses, we uncovered several active MITEs and their transposase sources [20].
Transposing from the research laboratory to the undergraduate classroom: The Dynamic Genome Courses (funded by the Howard Hughes Medical Institute Professor’s program)
Beginning in the fall 2011 quarter, Dr. Jim Burnette and I will be teaching a new course where incoming freshman will participate in cutting edge research derived from projects in my research laboratory. The Dynamic Genome will be the first course offered in the new Neil A. Campbell Science Learning Facility where students will study transposable elements in a state of the art classroom that includes computational and experimental laboratories. Please visit the course website.
References
- Fedoroff N, Wessler S, Shure M (1983) Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235-242.
- Wessler SR (1988) Phenotypic diversity mediated by the maize transposable elements Ac and Spm. Science 242: 399-405.
- Han Y, Burnette JM, Wessler SR (2009) TARGeT: a web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Research. pp. e78.
- Han Y, Wessler SR (2010) MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements (MITEs) from genomic sequences. Nucl Acids Res 38: e:199.
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596-1604.
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science. pp. 1112-1115.
- Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway MITEs. Genetics 163: 747-758.
- Zhang X, Jiang N, Feschotte C, Wessler SR (2004) PIF- and Pong-like transposable elements: distribution, evolution and relationship With Tourist-like miniature inverted-repeat transposable elements. Genetics 166: 971-986.
- Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR (2006) The transposable element landscape of the model legume Lotus japonicus. Genetics 174: 2215-2228.
- Zhang X, Wessler SR (2004) Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci 101: 5589-5594.
- Yang GJ, Weil CF, Wessler SR (2006) A rice TC1/mariner-like element transposes in yeast. Plant Cell 18: 2469-2478.
- Hancock CN, Zhang F, Wessler SR (2010) Transposition of the Tourist-MITE mPing in yeast: an assay that retains key features of catalysis by the class 2 PIF/Harbinger superfamily. Mobile DNA 1.
- Yang GJ, Zhang F, Hancock CN, Wessler SR (2007) Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci 104: 10962-10967.
- Bureau TE, Ronald PC, Wessler SR (1996) A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc Natl Acad Sci 93: 8524-8529.
- 15. Bureau TE, Wessler SR (1992) Tourist: a large family of inverted-repeat element frequently associated with maize genes. Plant Cell 4: 1283-1294.
- Bureau TE, Wessler SR (1994) Stowaway: a new family of inverted-repeat elements associated with genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6: 907-916.
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, et al. (2003) An active DNA transposon family in rice. Nature 421: 163-167.
- Naito K, Cho E, Yang GJ, Campbell MA, Yano K, et al. (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci 103: 17620-17625.
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, et al. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130-1134.
- Yang GJ, Nagel DH, Feschotte C, Hancock CN, Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325: 1391-1394.